| |
|
Research Projects
Given the that the ARP family have homologous zinc-dependent cytidine deaminase domains (CD) yet do not have common mRNAs or DNAs substrates, an important question to address is how do these enzymes distinguish where to deaminate cytidine or deoxycytidine. In fact failure to restrict ARP activity to the right nucleic acid target site, at the right time may give rise to mutational activity that could lead to a loss or gain of function and diseases such as cancer.
The Smith lab in collaboration with the laboratory of Joseph Wedekind (http://dbb.urmc.rochester.edu/labs/wedekind/Site/Wedekind_Home.html) are taking a structural approach to the question in trying to assess the active sites of these enzymes and their interaction with substrate. The identification of an orphan CDAR in yeast (CDD1) and its crystallization has enable us to model the three dimensional structure of mammalian APOBEC-1 and AID (Kefang, X., Sowden, M.P., Torelli, A.T., Dance, G.S.C., Smith, H.C. & Wedekind, J.E. (2004)
Structure of apoB Editing catalytic subunit 1 and activation induced deaminase predicted by the 2 Å resolution crystal structure of yeast RNA editing and cytidine deaminase CDD1. P.N.A.S. USA 101:8114-8119). This new model differs substantially from earlier predictions of the structure of APOBEC-1 in that it suggested that APOBEC-1 exists as a head to head dimer in which the linker region of one subunit completes the active site of the other subunit and visa versa (hence only dimers or higher oligomers are catalytically active), that the substrate binding pocket does not discriminate substrates based on ribose or deoxyribose binding but only can accommodate single stranded nucleic acids and that the linker region between the CD and PCD is flexible and unlike that found in cytidine or cytisine deaminase active on free nucleosides/nucleotides.
|
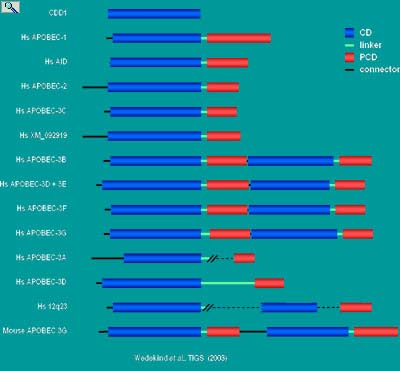
|
Schematic depiction of a structure-based alignment of APOBEC-1 in relation to its related proteins (ARPs). The gene duplication model for cytidine deaminases. CDD1 belongs to the tetrameric class of cytidine deaminases with a quaternary fold nearly identical to that of the tetrameric cytidine deaminase from B. subtilis . Such tetrameric enzymes exhibit the classical abbababb topology of the Zinc Dependent Deaminase Domain (ZDD) observed first in the Catalytic Domain (CD) of the dimeric enzyme from E. coli. According to the gene duplication model, an ancestral CDD1-like monomer duplicated and fused to produce a bipartite monomer. Over time a C-terminal Pseudo-Catalytic Domain (PCD) arose that lost substrate and Zn2+ binding abilities. The interdomain CD-PCD junction has been proposed to be flexible linker that forms part of the catalytic domain and facilitate or even determine substrate binding and selectivity. The putative function of the PCD is to stabilize the hydrophobic monomer core and to engage in auxiliary factor binding.
|
|
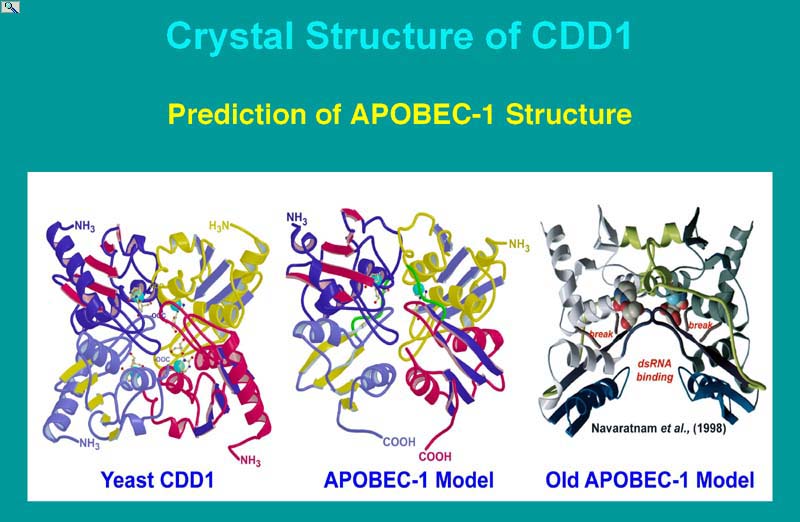
The structure of yeast CDD1 , yeast cytosine deaminase, E. coli cytidine deaminase and B. subtilis cytidine deaminase comparative modeling of HsAPOBEC-1 and HsAID, and the preparation of a structural template revealed major differences exist in the modes of substrate binding by enzymes that bind nucleosides and nucleotides (also RNA and DNA) from those that bind free pyrimidine bases. Comparative modeling of APOBEC-1-related proteins suggested that the domain interface between subunits influences the positioning of ribose binding loops as in cytidine deaminases and that a variably sized 'flap' at the catalytic site regulates access based on substrate size, which was corroborated in deaminase and RNA editing functional assays. The recent discovery of the requirement for AID in immunoglobulin gene rearrangements (see earlier discussion), as well as APOBEC-3G (CEM15) in the suppression of viral infectivity, are being pursued by the Smith lab in collaboration with the labs of Andrea Bottaro, Steve Dewhurst, Beak Kim (Dept. Microbiology and Immunology) and Joseph Wedekind (Dept. biochemistry and Biophysics) in terms of the question of substrates binding and specificity, and the role of auxiliary proteins in regulating activity. Although much information is known for APOBEC-1, making it a model system, different paradigms have been proposed for AID in which either DNA or RNA represents the primary substrate. Both AID and APOBEC-1 targeted DNA in E. coli. However, APOBEC-1 did not substitute for AID in immunoglobulin gene rearrangement, and AID could not substitute for APOBEC-1 in apoB mRNA editing, suggesting each enzyme has an inherent specificity for its own substrate. The models of APOBEC-1 and AID presented here indicated either DNA or RNA substrates could be accommodated by their active sites, but only in single-stranded form. Given these observations we remain open to both DNA-deamination and RNA-editing mechanisms as possible outcomes of all ARP biological activities.
(Kefang, X., et al. (2004) Structure of apoB Editing catalytic subunit 1 and activation induced deaminase predicted by the 2 Å resolution crystal structure of yeast RNA editing and cytidine deaminase CDD1. P.N.A.S. USA 101:8114-8119 and supporting information published with this article on the PNAS webpage ).
|
|
|